Have scientists finally solved tooth decay?
How new research on some of the worst dental bacteria could lead to exciting discoveries.
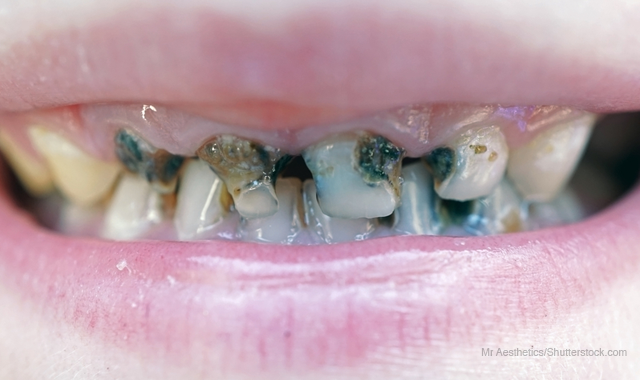
The cause of tooth decay has long been known - two kinds of bacteria (Streptococcus mutans and Streptococcus sobrinus) create a biofilm and acid that damages teeth.
Those two bacteria are often linked together by researchers in a group simply called mutans streptococci, since they both work together to form dental caries. However, much more is known about S. mutans, since it was first sequenced in 2002. S. mutans is more prevalent than its caries-causing twin and is more stable, making it much easier to study.
Until recently, very little was known about S. sobrinus. That changed when a group at the University of Illinois Bioengineering department sequenced the genomes of three strains S. sobrinus.
More emerging research: Are elderly patients with caries at risk of developing respiratory problems?
According to team leader Dr. Paul Jensen, an assistant professor at the university, the reason S. sobrinus is so little studied is that it is difficult to work with in the lab and it isn’t present in all people. However, it was known that S. sobrinus has a significant role in dental decay.
"Although it is rare, S. sobrinus produces acid more quickly and is associated with the poorest clinical outcomes, especially among children,” says Jensen. “If S. sobrinus is present along with S. mutans, you’re at risk for rampant tooth decay, which means there's some level of communication or synergy between the two that we don't understand yet."

Armed with this new knowledge about the S. sobrinus's genome, Jensen and his team are now building models that will allow them to study the relationship between the two bacteria and how they can be neutralized most effectively. The work is funded with a $218,000 grant from the National Institute of Dental and Craniofacial Research (NIDCR).
For instance, the researchers have already noted that the S. sobrinus doesn’t have what is known as quorum sensing, an ability many bacteria have that allows them to sense and react to other nearby bacteria. S. mutans has this ability in the form of peptides that reach out sense other S. mutans bacteria. Then, once a certain threshold is reached, the bacteria attack. Jensen and his team are trying to figure out why this isn’t the case for S. sobrinus.
“This is groundbreaking work because the field was plagued by a lack of information," says Jensen. "In 2018, it is surprising that we had a whole species [of bacteria] that causes disease and no complete genome of it.”
Jensen also notes that his team is working on a genomic-scale model, which will allow them to run virtually any experiment they want. With this model, he says, they could simulate what would happen if certain genes were deleted or if the bacteria were fed different sugars. Armed with this knowledge, his team will then make recommendations of drug targets for further experimentation. These therapies could eventually be used to lessen the destructive power of the bacteria.
Related article: Study finds link between oral bacteria and increased risk of esophageal cancer
“A lot of scientists are interested in S. sobrinus and they want to see what we find out,” says Jensen. “Using computation is allowing us to rationally demonstrate that this is the way to study S. sobrinus even though other lab-based experiments failed in the past.”
The team’s research on genome sequencing was published in Microbial Resource Announcements.
Henry Schein Now Exclusive Distributor of Curodont Remineralization Solution to DSOs
September 19th 2024The company announced it has reached an agreement with vVARDIS to serve as the exclusive distributor of their drill-free Curodont™ Repair Fluoride Plus product to larger dental service organizations in the U.S.
University of Texas Health Science Center San Antonio to Launch Center for Regenerative Sciences
June 17th 2024The center aims to translate preclinical discoveries into therapies for dental and craniofacial diseases, leveraging the school’s expertise in stem cell-based treatments and 3D printing technologies.